How I check the data
Check data to find misprints

Actually I analyse data from a thesis which measure urbanisation’s influence on physicochemical characteristics. The PhD student sample water once by month during 2017 at seven spots around a lake.
I think it is a nice occasion to explain how I check the data when I received them.
There are outliers?
First step: Outliers and Boxplot
Before to start anything, I test the data to find outliers (observation that has a relatively large or small value compared to the majority of observations Zuur et al. (2010)).
I start by draw boxplots by month (one by characteristic).
library(ggplot2)
for (i in 3:6)
{
bp<-ggplot(X) + geom_boxplot(aes(x = as.factor(Date), y = X[,i])) + geom_jitter(aes(x = as.factor(Date), y = X[,i]), col = X$Site, alpha = 0.2) + theme_minimal() + xlab("Date") + ylab(variable.names(X)[i])
print(bp)
}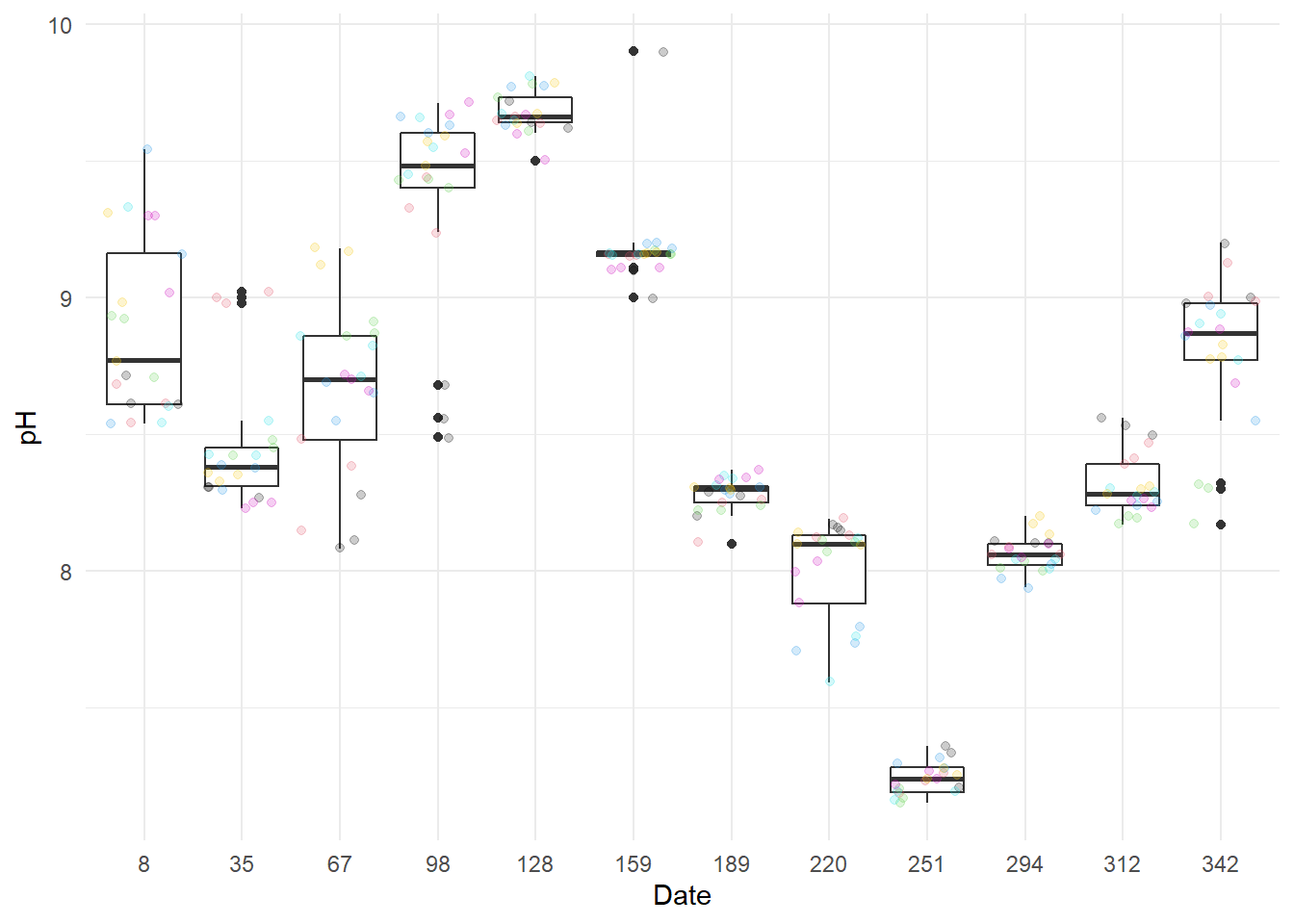
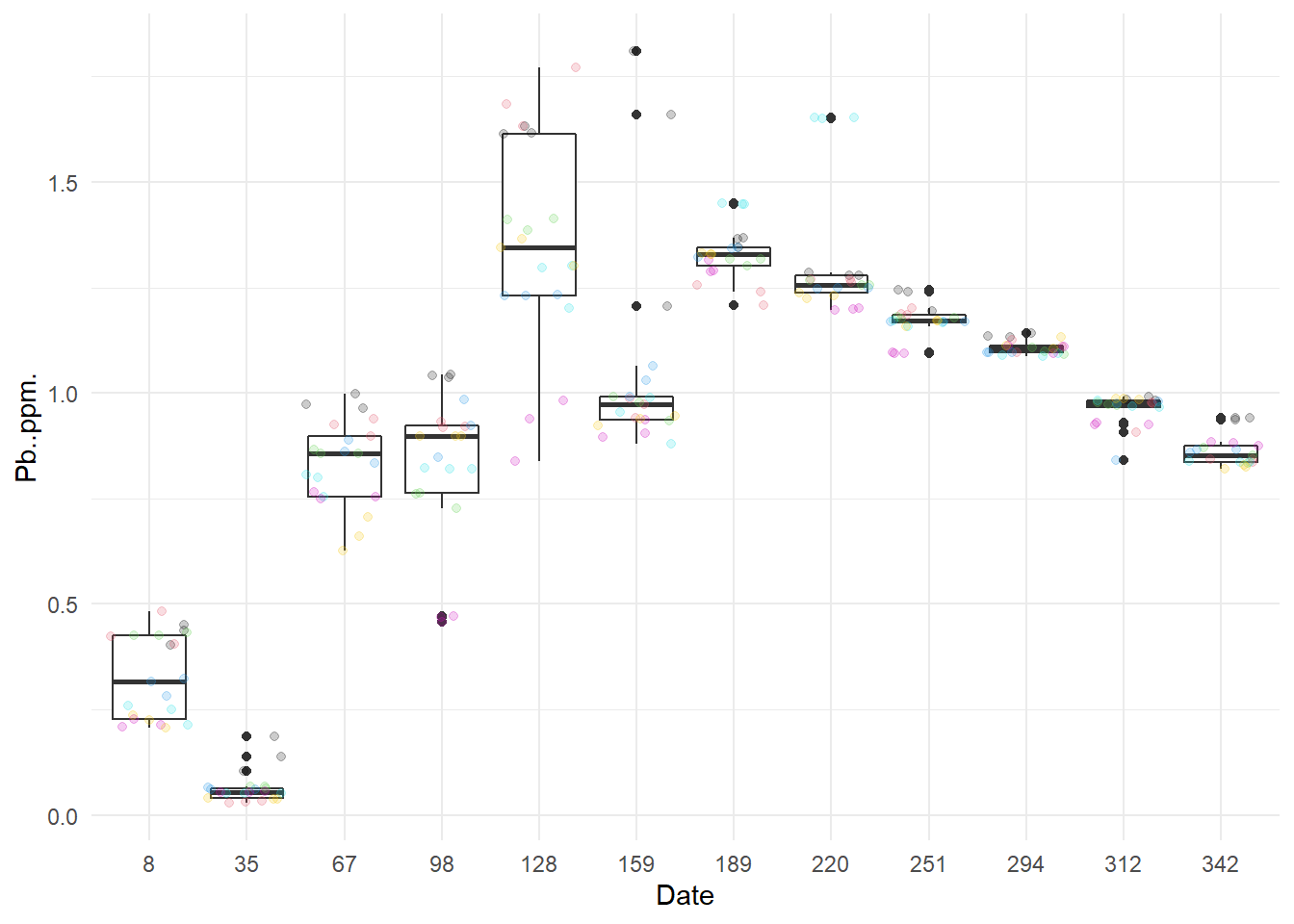
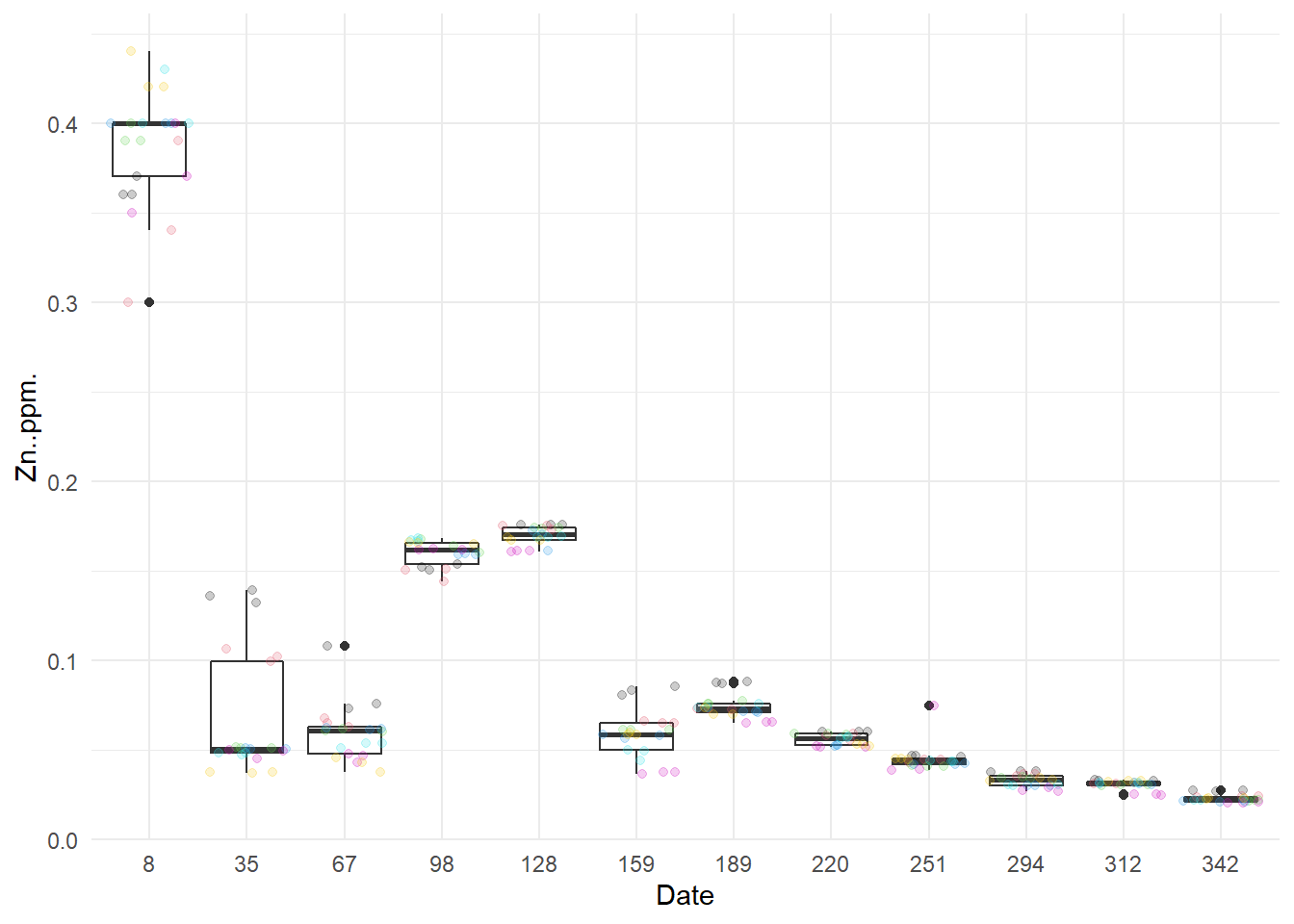
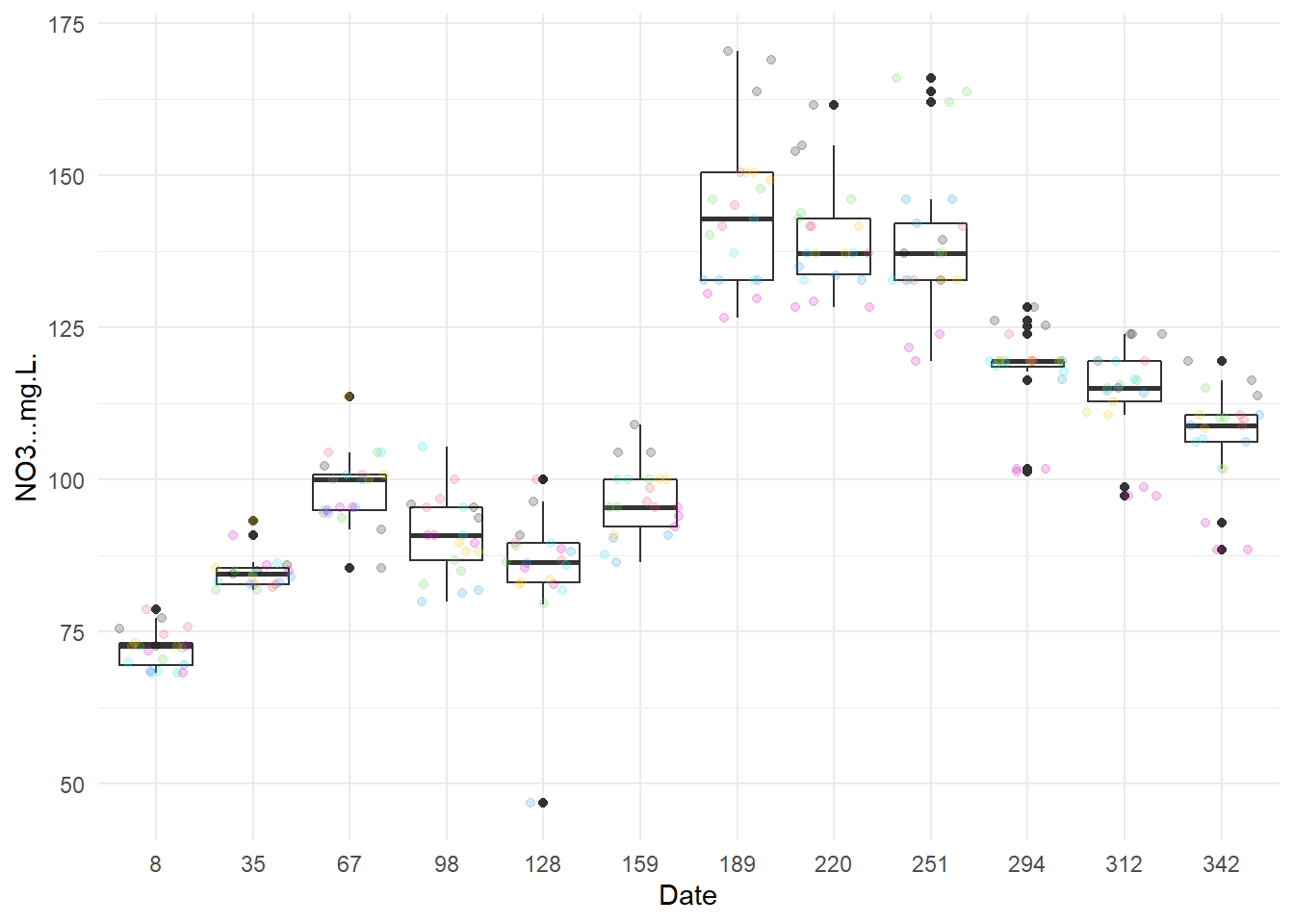 Except for NO3- at 128 days after the start of the year, there does not seem to have any outlier.
Except for NO3- at 128 days after the start of the year, there does not seem to have any outlier.
Second step: Outliers, means and standard error
In the data used, measurements were did three times each, so I calculated means and standard error with plotrix library.
library(plotrix)
X$rep<-paste(as.factor(X$Site), as.factor(X$Date))
# I create a variable to work on the repetition of measurements
# I create also the new database Z to have means and standard error according to spot and date
Site<-rep(1:7, each = 12)
Date<-rep(c(128,159,189,220,251,294,312,342,35,67,8,98), 7)
Z<-cbind(Site,Date)
CN<-colnames(Z)
for (i in 3:6)
{
M<-tapply(X[,i], X$rep, mean, na.rm=TRUE)
S<-tapply(X[,i], X$rep, std.error, na.rm=TRUE)
Z<-cbind(Z, M, S)
CN<-c(CN, paste(variable.names(X)[i],"Mean"),paste(variable.names(X)[i],"Se"))
}
colnames(Z)<-CN
write.table(Z, row.names=FALSE, "Physicochimical.txt", sep=";")
# Exporte table to work on it laterThen I analyse standard error by drawing.
Z<-read.table("Physicochimical.txt", header = TRUE, sep = ";")
o <- order(Z$Date)
Z<-Z[o,]
Z$Site<-as.factor(Z$Site)
for (i in c(4,6,8,10))
{
se<-ggplot(Z) + geom_point(aes(x = Date, y = Z[,i], color = Site)) + theme_minimal() + xlab("Date") + ylab(variable.names(Z)[i])
print(se)
}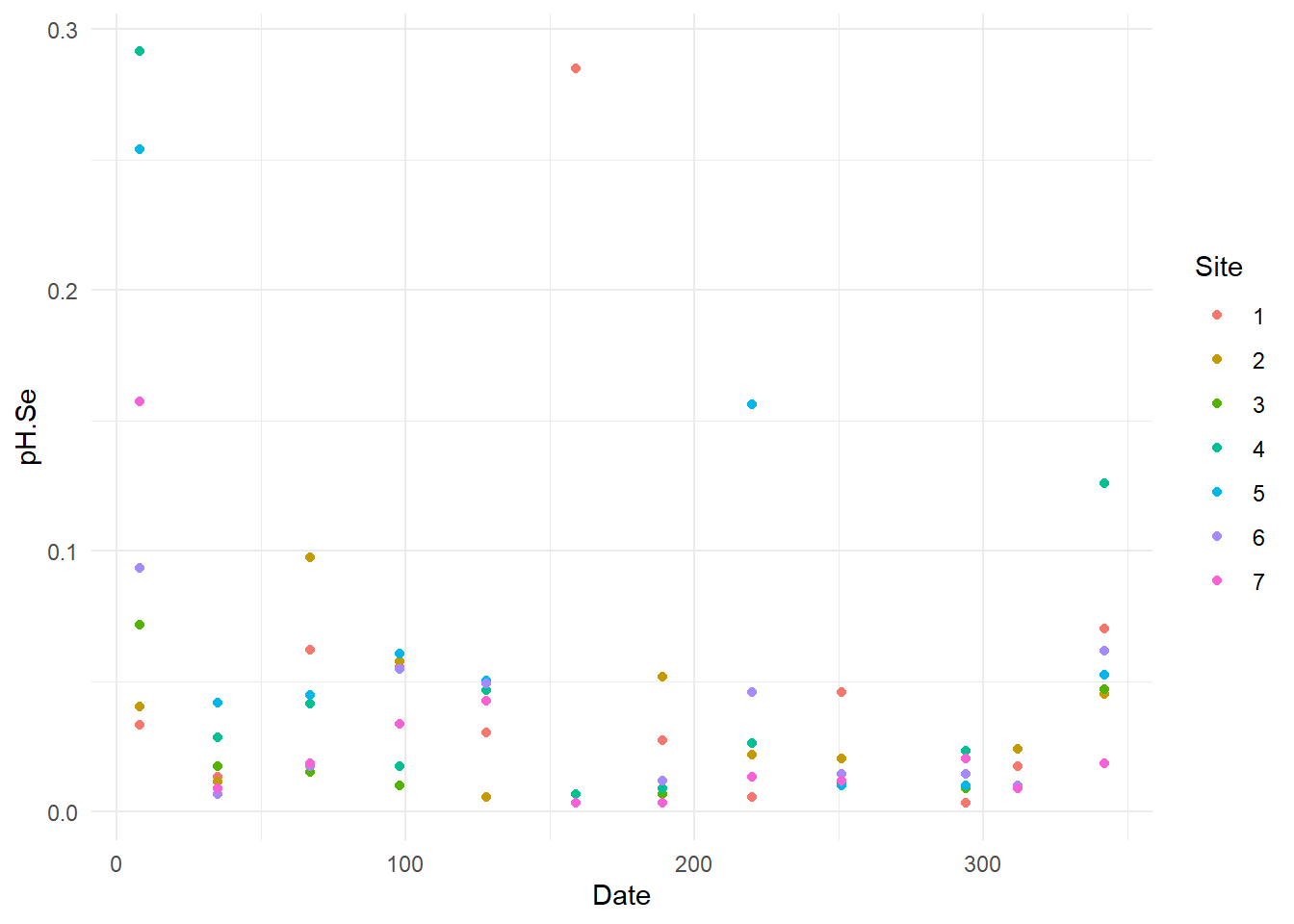
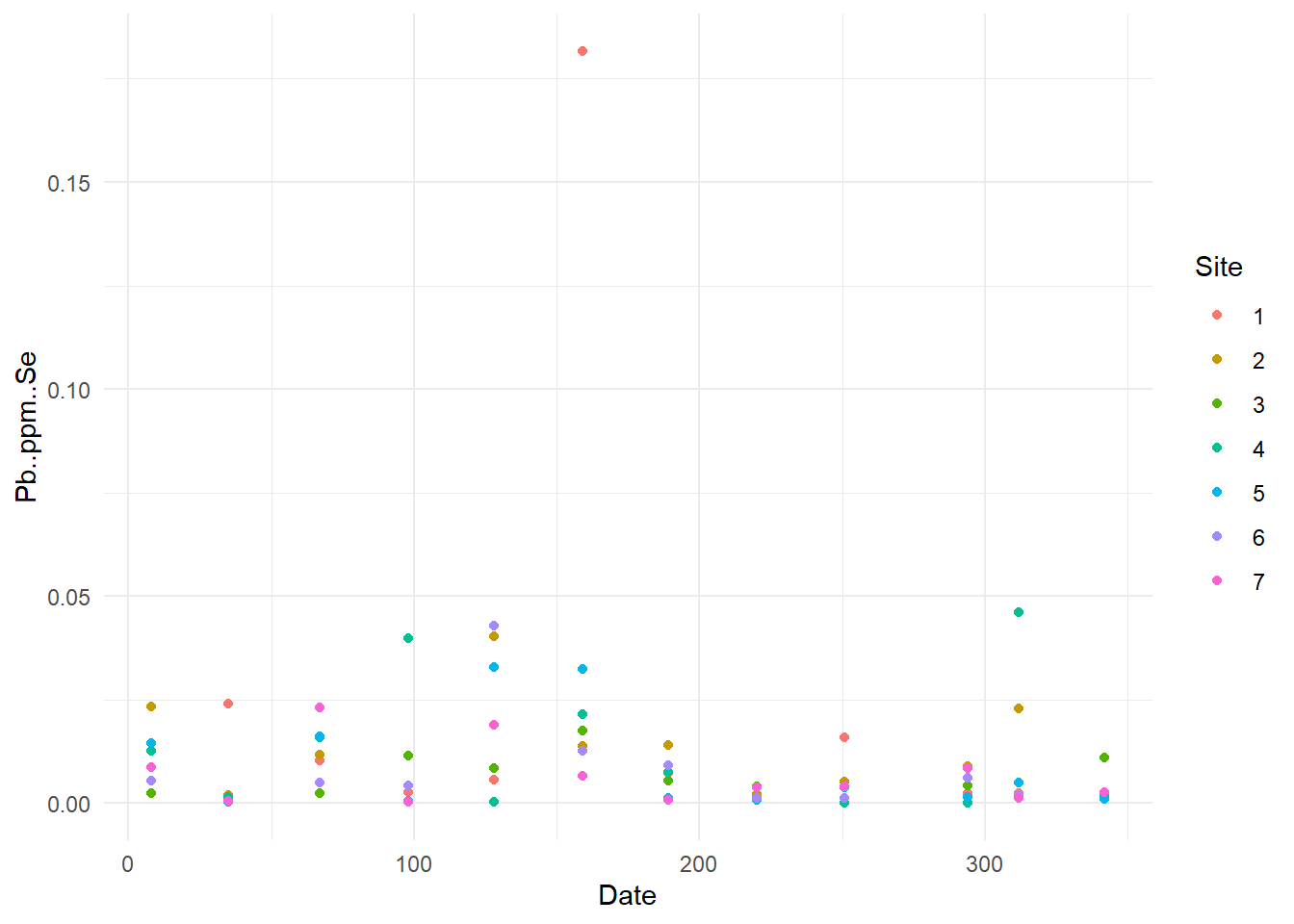

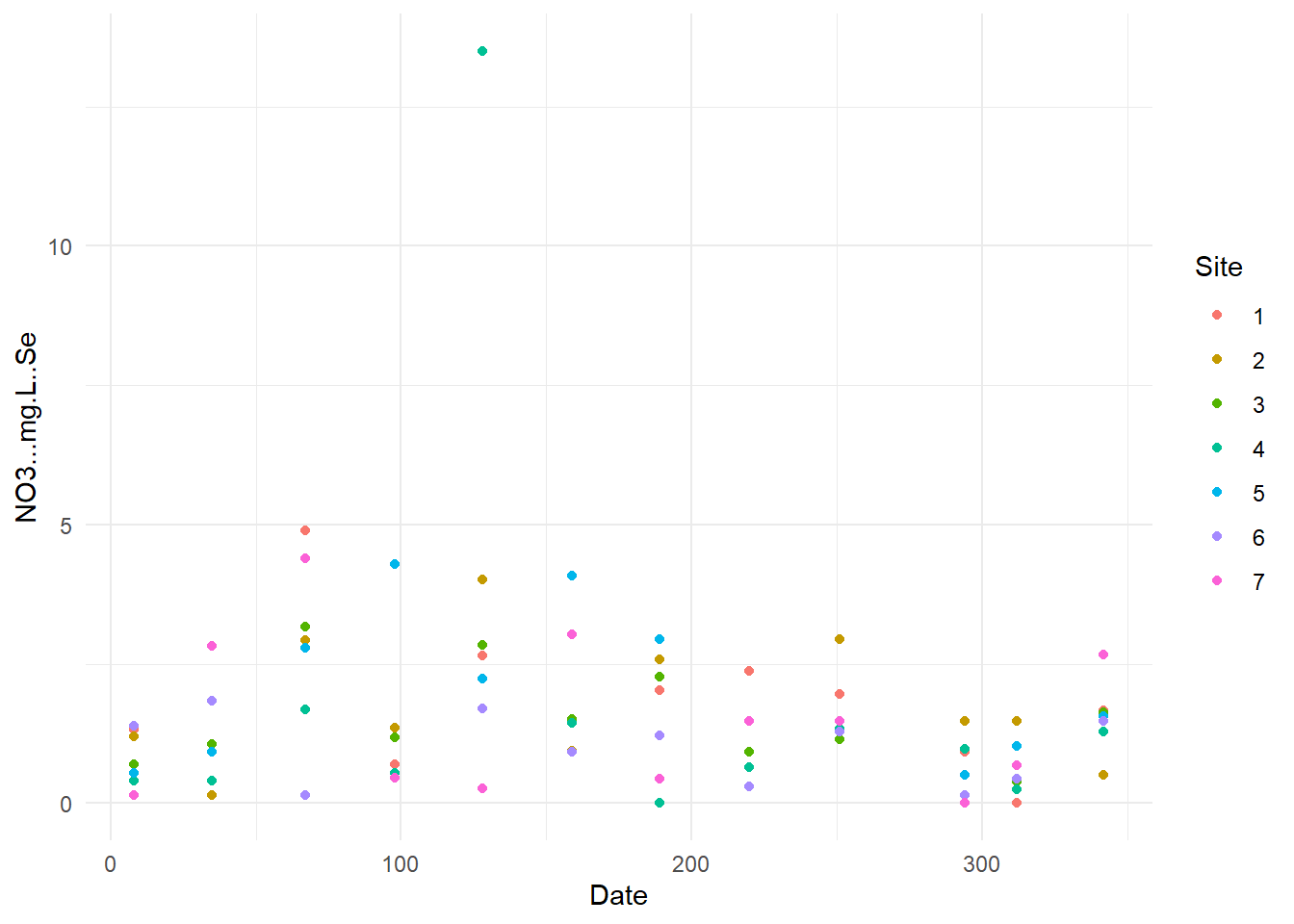 Now, we can see some standard error were higher than other (example for Pb one point has error higher than 0.1), so I will verify these specific data.
Now, we can see some standard error were higher than other (example for Pb one point has error higher than 0.1), so I will verify these specific data.
This method allows me to found and correct five misprint in the data. I will take in mind other outlier to check their influences on the futur analyses.
Verify the repeatability of the data
As in the previous blog article, I verify the repeatability of the corrected data with simple linear model.
Y$rep<-paste(as.factor(Y$Site), as.factor(Y$Date))
# The repeatability of the pH for exemple
mod<-lm(Y[,4]~Y$rep)
anova(mod)## Analysis of Variance Table
##
## Response: Y[, 4]
## Df Sum Sq Mean Sq F value Pr(>F)
## Y$rep 83 40.866 0.49236 735.84 < 2.2e-16 ***
## Residuals 168 0.112 0.00067
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Next time, I will describe how I do data exploration and first analyse.




Share this post
Twitter
Google+
Facebook
Reddit
LinkedIn
StumbleUpon
Pinterest
Email